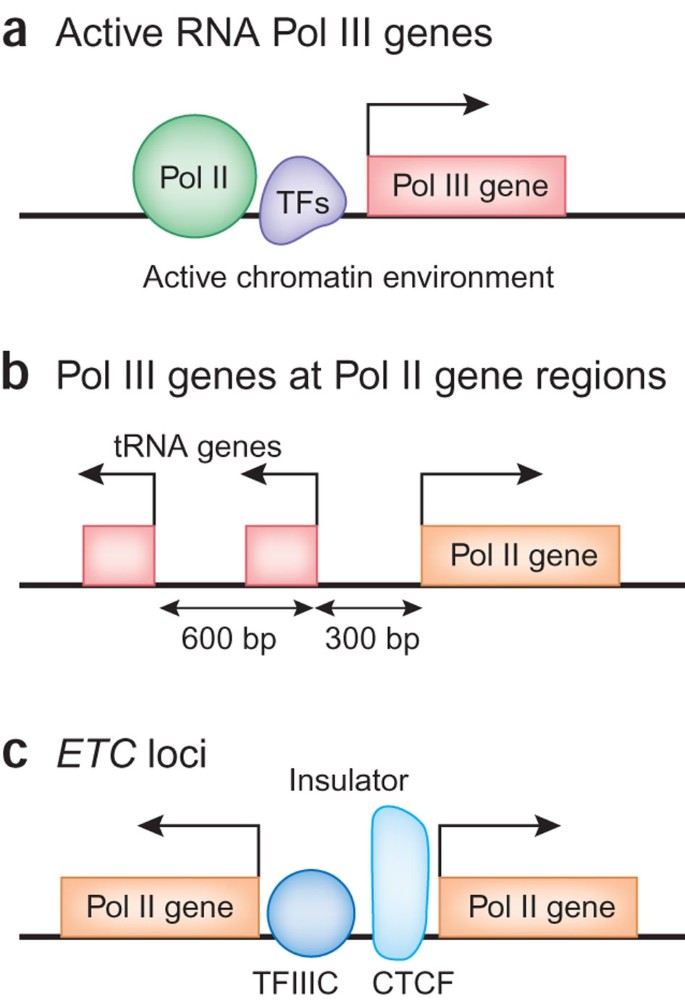
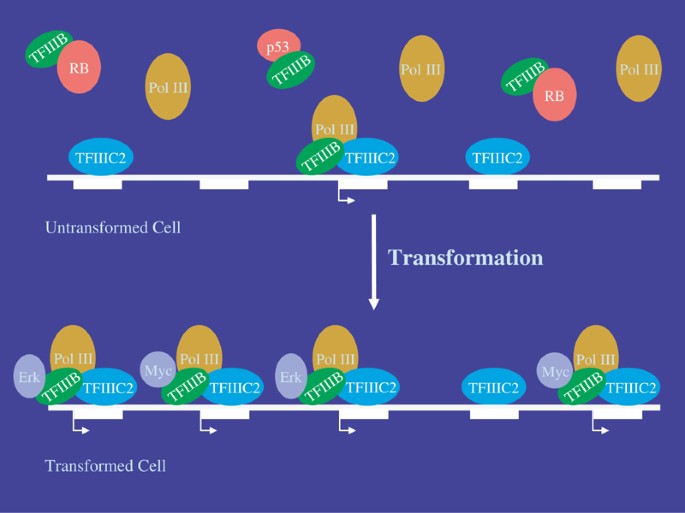
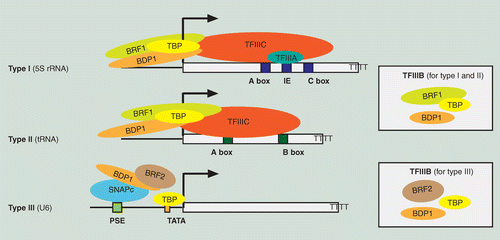
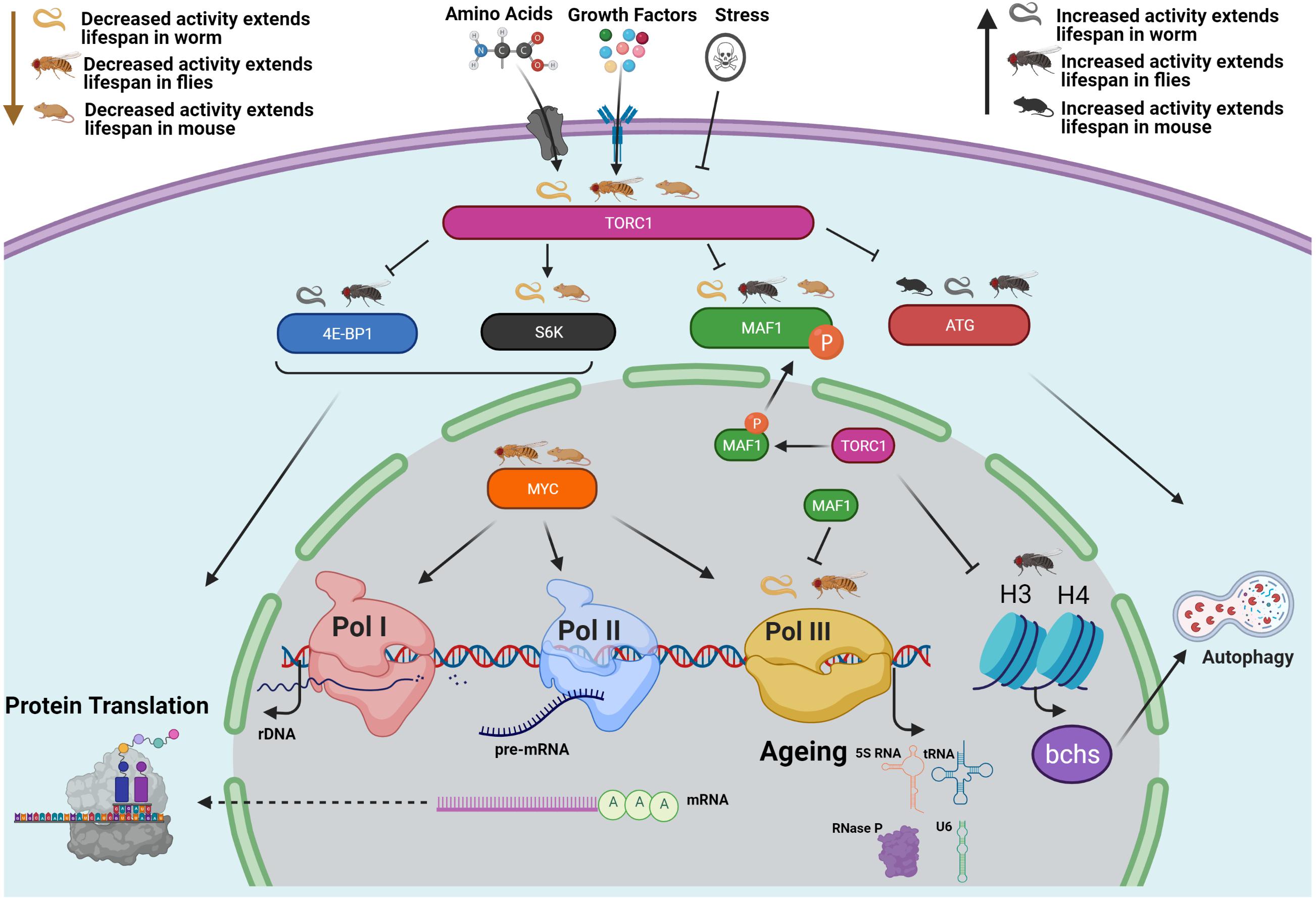
Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors | Nature Structural & Molecular Biology

DNA Polymerase in Prokaryotes and their mechanism of action( DNA Pol I ,DNA Pol II and DNA Pol III) - YouTube

Structural models of DNA polymerase III in polymerization and editing... | Download Scientific Diagram

DIF) Difference between Pol II and Pol III transcripts of Alu. In red... | Download Scientific Diagram

Researchers determine Pol III “identity” as important regulatory mechanism and likely disease factor in cancer | School of Molecular & Cellular Biology | UIUC

Devoted to the lagging strand—the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly | The EMBO Journal
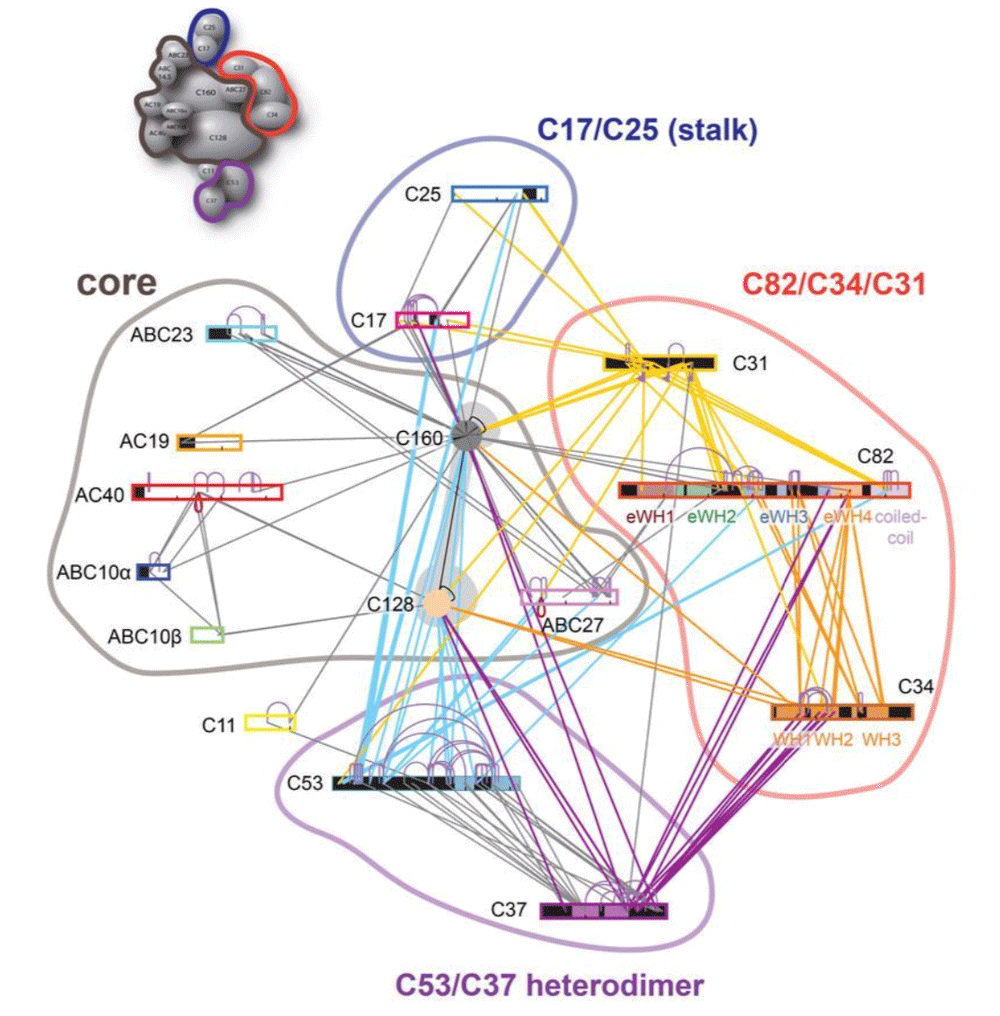
Integrative modelling of the apo RNA-Polymerase-III complex from MS cross-linking and cryo-EM data – Bonvin Lab